- Home
- News
- Spotlight on Science
- Structural basis...
Structural basis of an unusual carbocyclisation in nogalamycin biosynthesis
16-08-2016
The final steps in the biosynthesis of nogalamycin involve two atypical reactions, an epimerisation and a chemically challenging C–C-bond formation, which so far have been poorly understood. We show that these reactions are catalysed by two evolutionary-related non-heme iron α-ketoglutarate dependent enzymes. A limited set of amino acid changes in the active sites is responsible for the switch in chemistry. The two enzymes provide particularly striking examples as to how markedly different chemistry can evolve within an enzyme family.
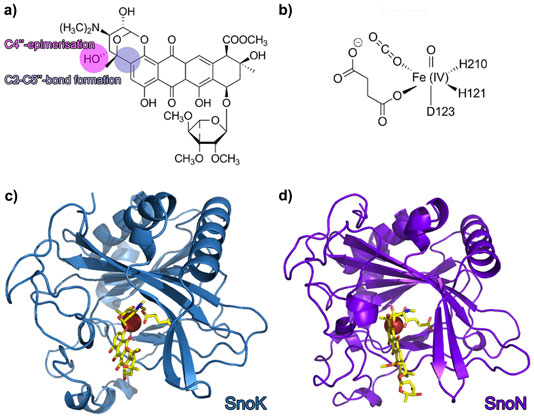
Over the last decade, research on natural product biosynthetic pathways in microorganisms and plants has revealed novel, unexpected chemistry and enzymatic mechanisms. One such example is the biosynthesis of the aromatic polyketide nogalamycin, produced by the soil bacterium Streptomyces nogalater. Like all glycosylated anthracyclines, nogalamycin is a cytotoxic metabolite and has generated some interest as a potential anti-cancer drug. Nogalamycin contains an unusual covalent attachment of one of the carbohydrate units to the aromatic ring system via a carbon-carbon linkage (Figure 1a). The formation of this bond during biosynthesis requires activation of an aliphatic C-H bond and reaction with an aromatic ring system. In spite of intensive studies into the biosynthesis, the mechanism of this carbocyclisation step has remained enigmatic. Previous studies also suggested that the final steps of nogalamycin biosynthesis involve an epimerisation converting the rhodosamine sugar moiety to nogalamine [1]. We have used genetics, biochemistry and protein crystallography to identify the enzymes responsible for these biosynthetic reactions and to characterise their hitherto unknown mechanisms. We discovered two homologous Fe(II) and α-ketoglutarate dependent enzymes (Figure 1b), denoted SnoN and SnoK, which catalyse these two key steps in the formation of nogalamycin. SnoK is responsible for the enzymatic carbocyclisation, while SnoN is an epimerase. The three-dimensional structures of SnoK (Figure 1c) and SnoN (Figure 1d) and several complexes with substrates were determined by protein crystallography, using data collected at beamlines ID29, ID30A-1 and ID30A-3.
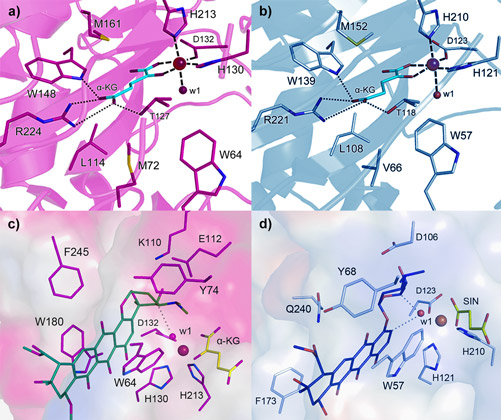
Both enzymes display the fold characteristic for the α-ketoglutarate-dependent oxygenase family, with the mononuclear iron ion located in the interior of the β-barrel fold. The co-substrate α-ketoglutarate is bound at the bottom of the channel and the environment of this sub-region is very similar in the two proteins (Figure 2a and 2b). The polyketide substrate is bound in an open cleft, lined with predominantly hydrophobic amino acids. Our biochemical studies and the crystal structures of the enzyme-substrate complexes point towards radical mechanisms involving a high-valent Fe(IV)=O species. The structural studies further imply that the initial catalytic steps required for activation of molecular oxygen are likely to be identical in SnoN and SnoK and occur in a manner similar to other members of the protein family. The exceptional feature of SnoK is the use of the ferryl-oxo species for an oxidative carbocyclisation reaction. Such chemical transformations are challenging to accomplish selectively in organic synthesis. In SnoK this reaction is initiated by the Fe(IV)=O species which abstracts a hydrogen atom from the primary substrate leading to the generation of a substrate radical. The intramolecular ring closure is then accomplished after attack of the aromatic C2 carbon atom on the radical center at C5´´.
In spite of the different reactions catalysed by these enzymes, they display significant amino acid sequence identity (46%) and a high degree of overall structure similarity. Major differences between the enzymes appear to be restricted to parts of the active site cleft involved in recognition and positioning of the polyketide substrates. These variations position the reactive atoms differently in front of the Fe(IV)=O centre and are likely to be critical for the differences in catalysis in the two enzymes. In SnoN (Figure 2c), the substrate is bound such that the site of stereoinversion is close to the iron centre. In SnoK (Figure 2d), the C2 carbon atom of the anthracycline aglycone and C5¢¢ of the carbohydrate are positioned adjacent to the metal centre facilitating carbocyclisation.
The characterisation of SnoN and SnoK presents an illustrative example of divergent enzyme evolution and provide a compelling example of how subtle differences in positioning of substrates may lead to greatly different chemistry in homologous enzymes. Furthermore our findings expand the repertoire of reactions reported for this enzyme family.
Principal publication and authors
Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway, V. Siitonen (a), B. Selvaraj (b), L. Niiranen (a), Y. Lindqvist (b), G. Schneider (b) and M. Metsä-Ketelä (a), PNAS 113, 5251-5256 (2016); doi: 10.1073/pnas.1525034113.
(a) Department of Biochemistry, University of Turku (Finland)
(b) Department of Medical Biochemistry & Biophysics, Karolinska Institutet, Stockholm (Sweden)
References
[1] V. Siitonen, M. Claesson, P. Patrikainen, M. Aromaa, P. Mäntsälä, G. Schneider, M. Metsä-Ketelä, ChemBioChem 13, 120–128 (2012).
Top image: The structure of SnoK, one of the key enzymes in the biosynthesis of nogalamycin, a potential anti-cancer drug.